Overview
In a multicenter study, our ODAP algorithm allows researchers to fit a Poisson regression model across multiple data sets, without the need of directly sharing subject-level information. The Poisson regression model is a commonly used approach to model a count outcome and a set of explanatory variables, which is a foundational method for studying associations, causal effects, and predicting frequency of a certain event. In addition to traditional Poisson regression, ODAP also allows for Quasi-Poisson regression, resulting in standard errors corrected for overdispersion. Before conducting the multicenter analysis, all datasets have to be converted to a unified format where all the variables are defined in a standard way (e.g, OMOP Common Data Model). We require one of the sites to be the coordinating site, and the rest to be the participating sites. The coordinating site is responsible for obtaining and broadcasting the initial value of model parameters, synthesizing information obtained from other sites, and obtaining the final results. The participating sites only need to calculate the aggregated data and transfer them to the coordinating site. A more detailed description is provided below.
Poisson Regression Model
Suppose we have K sites, and the coordinating site is the first site. For the i-th individual in the j-th site, we denote the count outcome to be \(y_{ij}\), and a vector of explanatory variables to be \(z_{ij}\). Let \(x_{ij} = (1, z_{ij}^{T})^T\) The Poisson regression model assumes that
\(P(Y_{i} = y_{i}) = \frac{e^{-\lambda_{i}} \lambda_{i}^{y_{i}}}{y_{i}!}\) with \(log(\lambda_{i}) = X_{i}^{T}\beta\)
where \(\beta\) is the vector of intercept and regression coefficients.
Algorithm
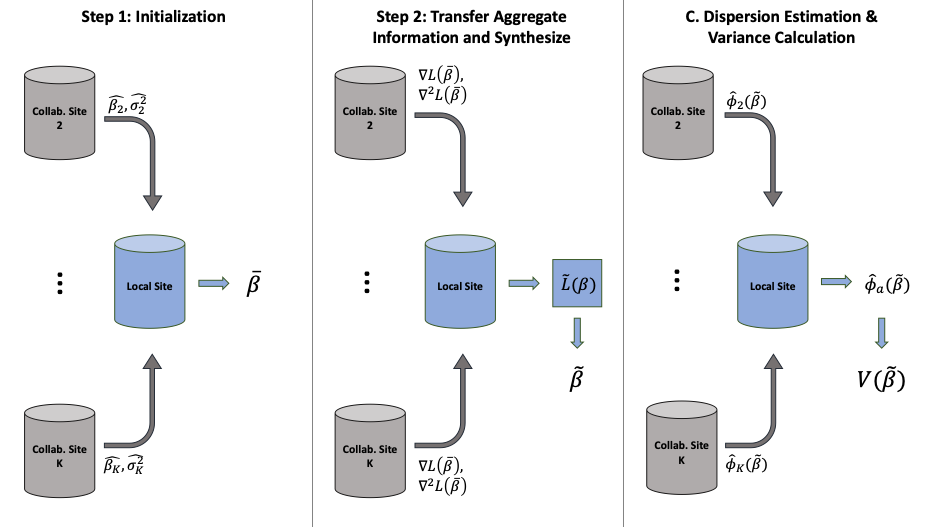
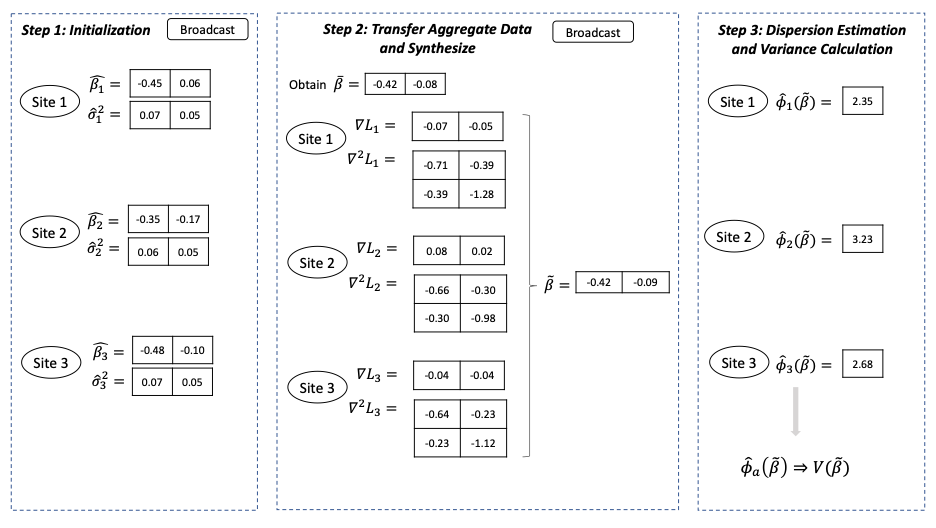
Our algorithm has three steps:
- First, each site fits a Poisson (or quasi-Poisson) regression model using its own data and sends point and variance estimates to the local site, where meta-analysis is done to compute initial estimates \(\bar{\beta}\). Initial estimates \(\bar{\beta}\) are then sent to all collaborating sites.
- Second, each collaborating site calculates first and second order gradients of their own likelihood function evaluated at the initial value \(\bar{\beta}\). These aggregate data are then transferred to the coordinating site, which combines these data with its own aggregate data to obtain \(\bar{\beta}\), an improved estimate of \(\beta\). If performing distributed quasi-Poisson regression, estimates \(\tilde{\beta}\) are then sent to all collaborating sites.
- (Optional, only if using quasi-Poisson regression) Third, each collaborating site estimates dispersion using \(\tilde{\beta}\), and these dispersion estimates are sent to the coordinating site where a weighted average is calculated. This weighted average, \(\hat{\phi}_{a}(\tilde{\beta}\)), is used to scale the variance of \(\tilde{\beta}\).
Figure 2 below gives an example of the information shared in each step in a setting with one explanatory variable in the model. For p explanatory variables, each site needs to transfer in total (p+1) + p* (p+1) numbers.
 Sample code
Sample code
Set the “control” in R as below to start ODAP algorithm
control <- list(project_name ='Lung cancer study',
step = 'initialize',
sites = c('site1', 'site2', 'site3'),
heterogeneity = FALSE,
model = 'ODAP',
family = 'poisson',
outcome = "CrabSatellites",
variables = c('width', 'weight'),
optim_maxit = 100,
lead_site = 'site1',
upload_date = as.character(Sys.time()) )
demo(ODAP)